Content
Search module
How to find the entry of all important information in the database by keywords?
Enter keywords to search for the corresponding information such as SRG regulatory network, homologous genes, expression dynamics across different conditions.
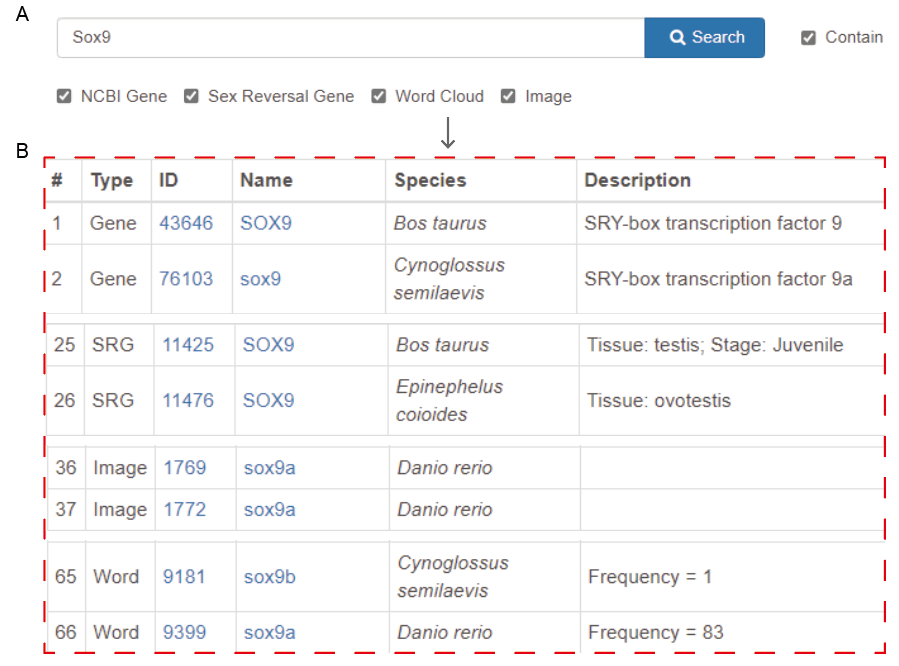
A. Enter the keyword
Keyword types:
- Symbol of gene, e.g. "SOX9".
- Alias of gene, e.g. "CMD1".
- Description of gene, e.g. "SRY-box transcription factor 9".
- Function of gene, e.g. "transcription factor".
B. Get the results
Result types:
- Gene: All the homologous genes annotated in NCBI.
- SRG: Sex reversal genes collected in the database.
- Word: Word cloud items collected in the database.
- Image: ISH Images collected in the database.
Searching for information in a specific section (e.g. GENE) will help filter the information.
Please uncheck “Contain” to achieve precise search.
Note: Although SRG, Word and Image do not have alias and description information, they can also be retrieved by the alias and description information of the corresponding gene.
Species module
How to get detailed information of Oreochromis niloticus in this database?
For the “SPECIES” module, the evolutionary tree constructed for the 18 sex reversal species is displayed in the main page. Users can click on any species to get detailed description, genome information and sex reversal inducements for this species.
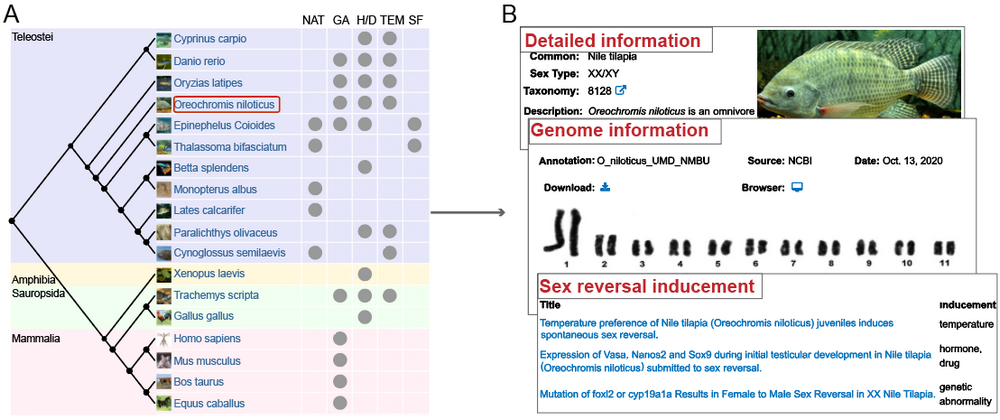
A. Click on your interested species. Evolutionary relationship and their sex reversal inducements of the 18 species belong to Teleostei, Mammalia, Sauropsida, and Amphibia are displayed in this page. NAT: Natural; GA: Genetic abnormality; H/D: Hormones or drugs; TEM: Temperature; SF: Social factors.
B. Get the results. Detailed description, genome information, and references related to sex reversal in each species are shown.
SRG module
How to search regulatory network for any SRG (Sex Reversal associated Gene)?
The “SRG” module includes word cloud, regulatory and PPI networks, and search pages. For any validated SRG, ASER allows users to obtain its regulators (including genes, hormones, drugs), targets, and the associated modes of regulation. At the same time, PPI network of these SRGs in different species are also displayed.
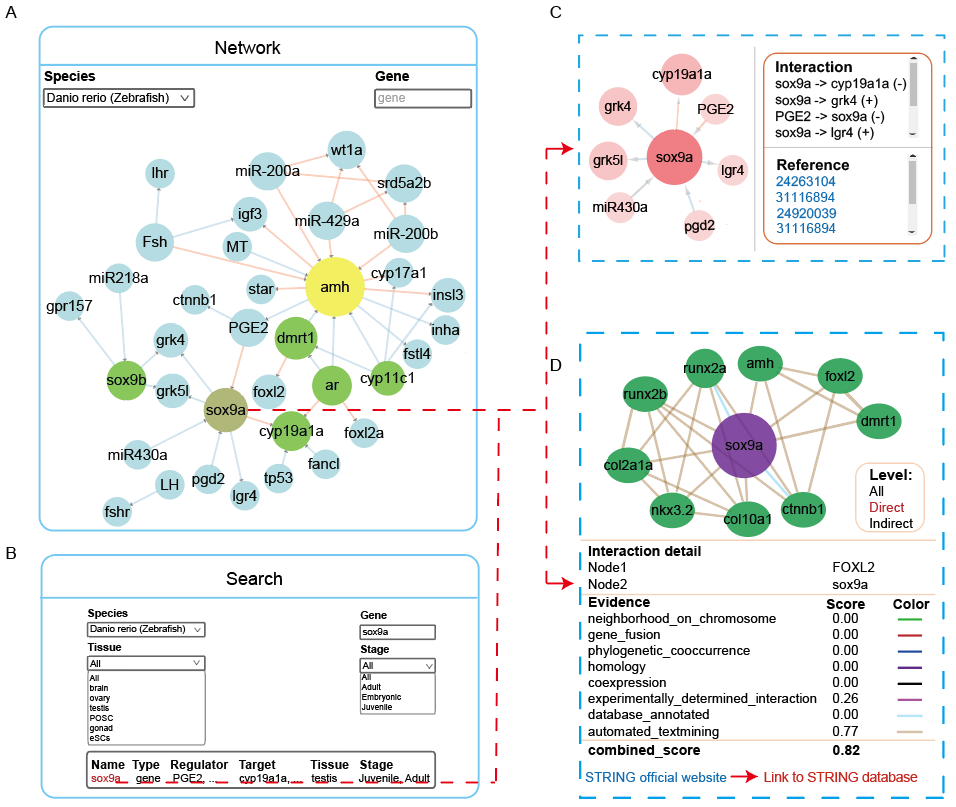
A. Select your interested species and click on a SRG to get detailed information in Network module.
B. Search a specific SRG to show its regulatory network and other information via more detailed constraints, such as tissue and developmental stage, and etc in Search module.
C. Regulatory network for a specific SRG and literature evidence for regulatory relationships are shown. The color of edges indicate regulatory relationship, bule: positive, red: negative.
D. Integrated Protein-Protein Interaction (PPI) network for selected SRG in different species. The colors of the edges are used to distinguish known interactions (experimentally_determined_interaction, database_annotated), predicted interactions (neighborhood_on_chromosome, gene_fusion, phylogenetic_cooccurrence), and other types (homology, coexpression, automated_textmining). The PPI network was collected from the STRING database for a quick comparison with the SRG network. More information is provided on the STRING official website.
We compiled a list of the most commonly used genes or drugs related to sex reversal and differentially expressed genes (female-specific:50, male-specific:50), then presented them as a word cloud. The word cloud figure is dynamically presented by species with hyperlinks on the nodes.
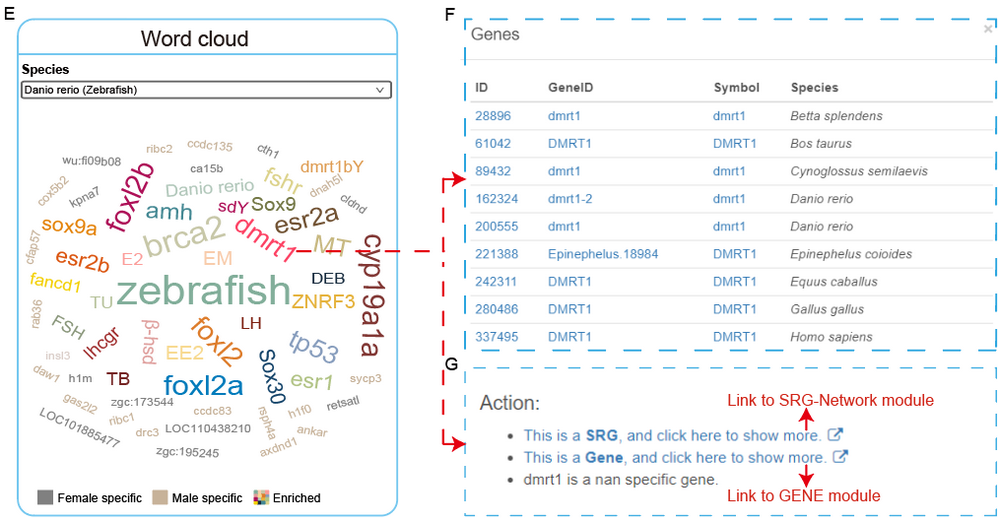
E. Select your interested species and click on a gene.
F. Cross-species information are shown in the pop-up window.
G. Links to SRG-Network module and GENE module to get detailed information such as regulatory network and expression information are provided. In addition, upstream references of selected word are shown in the website.
Gene module
How to obtain homologous genes and expression information of interested gene?
The “GENE” module provides different kinds of data for any annotated gene, some of which are linked to the “BROWSER” module for visualization. The links corresponding to the query gene are shown in the search page by species and gene symbol. The detailed information for a specific query gene includes its orthogroup in all species, predicted motifs, and similar known motifs. In addition, the gene expression quantifications in FPKM across different stages, tissues, and conditions are shown as bar plot and in details as table.
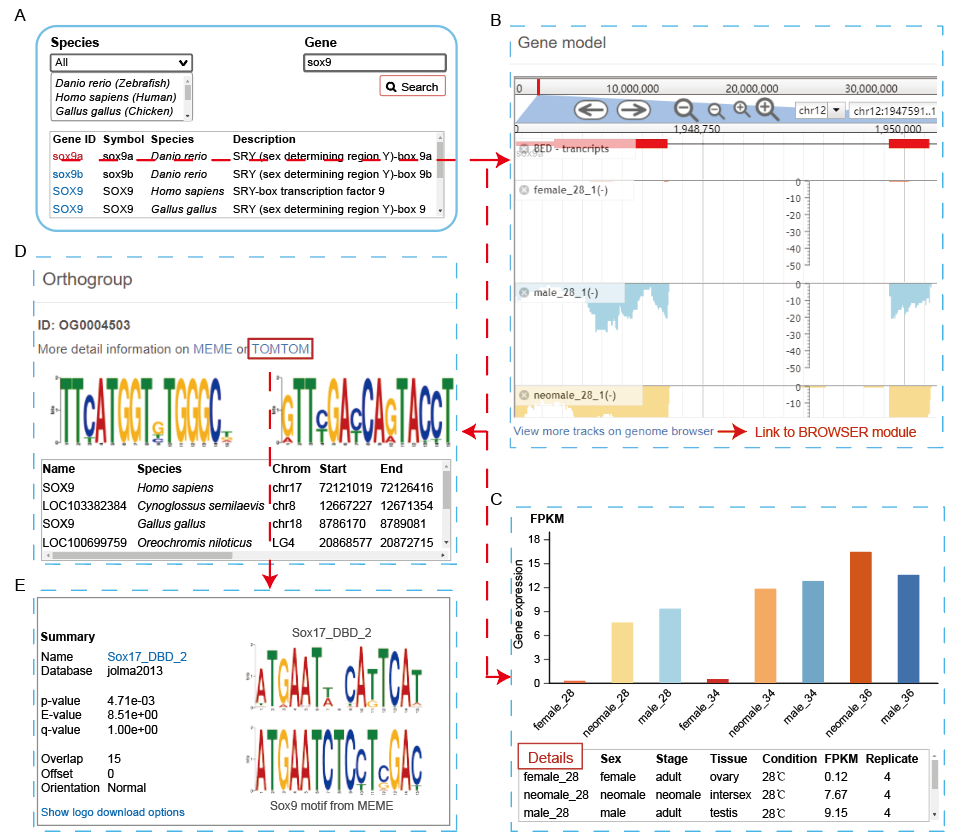
A. Select your interested species and gene.
B. Gene structure and representative expression track in different gender are shown. Link to the “BROWSER” module for visualization of more data is provided.
C. Gene expression quantifications in FPKM across different stages, tissues, and conditions are shown as bar plot and in details as table. User can specify different constraints (Sex,Stage and Tissue) to do fine search.
D. Homologous genes in the 18 species and motifs corresponding to the query gene are diaplayed.
E. The known motifs similar with conserved motif for selected gene are presented.
Image module
How to explore spatial expression of SRGS in the gonads?
We collected available (Fluorescence) in situ hybridization and immunocytochemistry (ISH, FISH and ICH) data related to SRGs. The images were classified by gene, differentiation status, developmental period, and gender. We manually added descriptions for those images based on the original figure legends and articles.
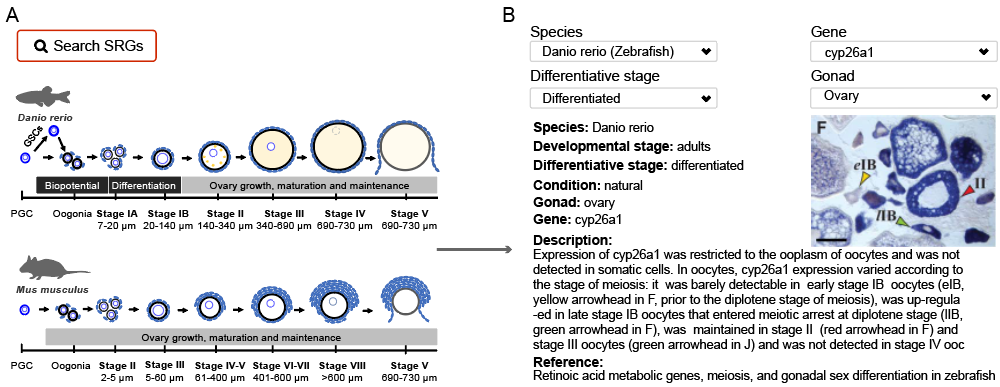
A. we summarized the morphological characteristics of zebrafish and mouse ovary at different developmental stages to help users better understand the content of this module.
B. Specify different constraints to do fine search.
Constraint types:
- Species, e.g. "Zebrafish".
- Gene, e.g. "cyp26a1".
- Differentiative stage, e.g. "differentiated”.
- Gonad, e.g. "Ovary".
Browser module
How to visualize gene expression dynamics during sex reversal in browser?
The RNA-seq signal profiles are displayed in BROWSER and the tracks can be customized easily including color, scale, height, and etc. The orthologue genes and 18-way conservation scores for the query gene can be inspected in BROWSER tracks. For any species, the available tracks can be dynamically selected or unselected.
Search by gene name, alias and function are achieved by two ways:
From “SEARCH” module.
- Enter interested gene.
- Click on blue gene name whose type is Gene.
- Click on “View more tracks on genome browser” in the Gene model.
From “GENE” module.
- Enter interested gene.
- Click on “View more tracks on genome browser” in the Gene model.
You can also search by genome location in “BROWSER” module as follows.
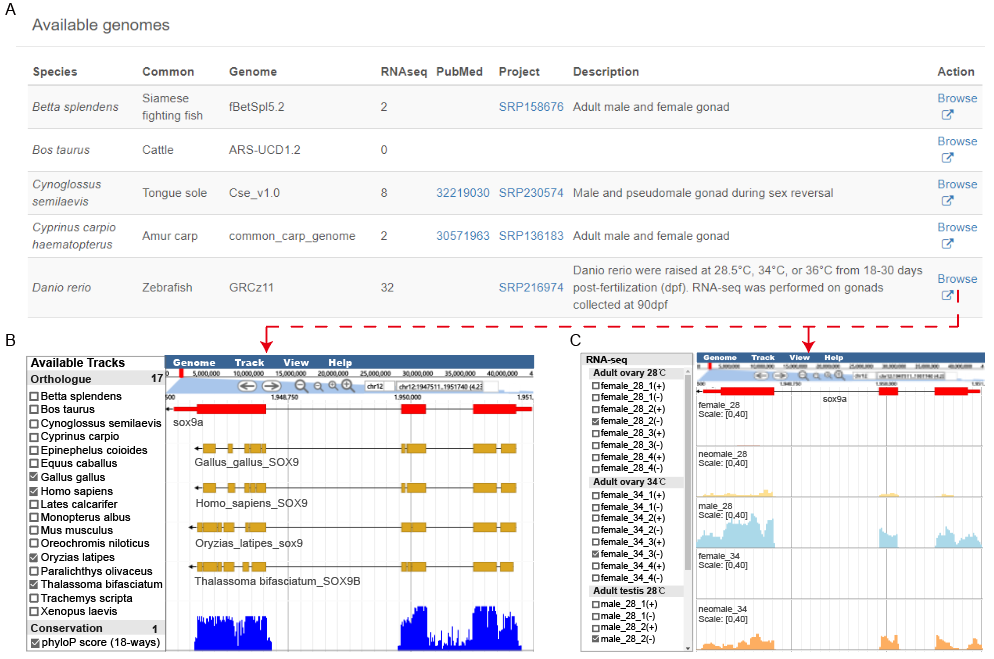
A. Detailed descriptions of available genome and data are shown.
B. Orthologue tracks and Conservation scores are displayed. For the orthologue tracks in a reference species, the CDS sequences of genes in other species were mapped to the reference genome. The yellow blocks represent high sequence identity region. Conservation scores of selected position across 18 species, High peaks represent high conservative scores.
C. The RNA-seq signal profiles of different stages, tissues, and conditions are displayed.
If you want to compare the tracks between different species, we provide a two-species interface in “BROWSER” module.
Tool module
How to use LiftOver?
LiftOver is a necessary step to bring all genetical analysis to the same reference build. LiftOver can have three use cases:
- Convert genome position from one genome assembly to another genome assembly. In most scenarios, we have known genome positions in NCBI build 36 (UCSC hg 18) and hope to lift them over to NCBI build 37 (UCSC hg19).
- Convert dbSNP rs number from one build to another.
- Convert both genome position and dbSNP rs number over different versions.
The usage of liftOver in the terminal is as follows:
liftOver oldFile map.chain newFile unMapped
In this command, oldFile is the uploaded file in bed format.
newFile is the file that successfully mapped and unMapped is the file that failed to be transformed.
The map.chain file has the old genome as the target and the new genome as the query.
On this interface, you should select your source assembly and target assembly, and then
upload the files to be converted. After submission, you will be able to download
the result files after successful conversion and the failed files.
For a more flexible conversion, you may download the chain file from the DOWNLOAD section and run the liftOver
on your local computer.
More detail about the liftOver, please visit LiftOver wiki.
Download module
What kind of data can be downloaded?
The structure of the available data is shown as below: (token Homo sapiens as an example)
-
Homo sapiens
-
Genome and genes
- genome.fasta
- genome.sizes
- annotation.gff.gz
- annotation.bed.gz
- chromosomes.tsv
- genes.tsv
-
SRG and network
- srg.tsv
- network.tsv
-
RNA-seq
- tracks (bigwig files)
- fpkm.tsv
-
liftOver
- query.chain
-
Genome and genes